The HIV virus infects roughly 40 million people worldwide; the retrovirus compromises the immune system, paving way for other diseases. While there are treatments for this disease, only about half of the people infected receive it, and a fraction of them also have side effects or reject the treatment, making it ineffective. A better understanding of the molecular mechanisms of different processes during the HIV replication cycle will benefit the development of new anti-viral therapeutics.
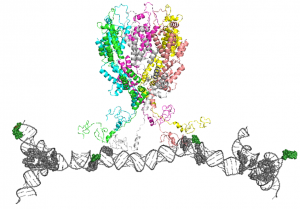
We are focused on studying how Gag can selectively package the viral dimeric genome among a large excess of other RNAs in the host cell cytosol. We propose that clustering of Gag on the 5’ UTR of viral genomic RNA (the packaging signal) promotes the formation of Gag hexamers, which will function as the nucleation site to recruit other Gag molecules for viral assembly. To validate this hypothesis, we seek to characterize the interactions between Gag and the packaging signal including the binding stoichiometry and binding affinity using electromobility shift assay and isothermal titration calorimetry. The formation of Gag hexamers are probed using chemical crosslinking and electron microscopy. Our ultimate goal is to solve the structure of this nucleation complex, which will reveal the detailed molecular mechanism for how Gag can selectively package the dimeric genome.
