Iron is involved in crucial biological processes and is essential for nearly all living organisms. Because different oxidation states of iron are dominant within various biological niches, bacteria necessitate oxidation-state specific iron uptake systems to meet their iron requirements. In anaerobic and/or acidic environments such as those in subgingival biofilms, the more soluble ferrous (Fe2+) form of iron is typically pervasive. Ferrous iron is transported via the Feo system, which contributes to the virulence of several pathogenic bacteria, including the common human pathogen Porphyromonas gingivalis. However, little is known about the mechanism of ferrous iron transport, which could be targeted as a potential way to combat antibiotic resistance.
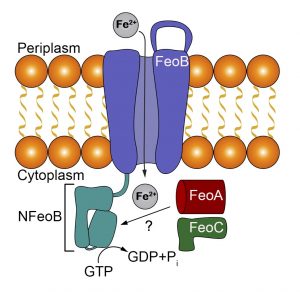 Within the Feo system there may be as many as three proteins: FeoA, FeoB, and FeoC. FeoA and FeoC are soluble accessory proteins, whereas FeoB is the main transporter and is a membrane protein. Previous experiments in our lab have shown that there is an interaction between FeoA and FeoB. Our current work aims to understand the atomic-level details of this interaction within the P. gingivalis pathogen. To do so, we are optimizing purification conditions of P. gingivalis NFeoAB, a naturally occurring fusion protein. Once optimized, we plan to crystallize this fusion in order to gain three-dimensional insight into how FeoA and FeoB interact. In parallel, we are actively pursuing expression and functional characterization of the intact close homolog Porphyromonas gulae FeoAB. These studies will provide insight into how FeoA and FeoB interact in one-component systems and may provide a future target for novel therapeutics to treat gingivitis.
Within the Feo system there may be as many as three proteins: FeoA, FeoB, and FeoC. FeoA and FeoC are soluble accessory proteins, whereas FeoB is the main transporter and is a membrane protein. Previous experiments in our lab have shown that there is an interaction between FeoA and FeoB. Our current work aims to understand the atomic-level details of this interaction within the P. gingivalis pathogen. To do so, we are optimizing purification conditions of P. gingivalis NFeoAB, a naturally occurring fusion protein. Once optimized, we plan to crystallize this fusion in order to gain three-dimensional insight into how FeoA and FeoB interact. In parallel, we are actively pursuing expression and functional characterization of the intact close homolog Porphyromonas gulae FeoAB. These studies will provide insight into how FeoA and FeoB interact in one-component systems and may provide a future target for novel therapeutics to treat gingivitis.